Publications
Highlights
(For a full list see below or go to Google Scholar)
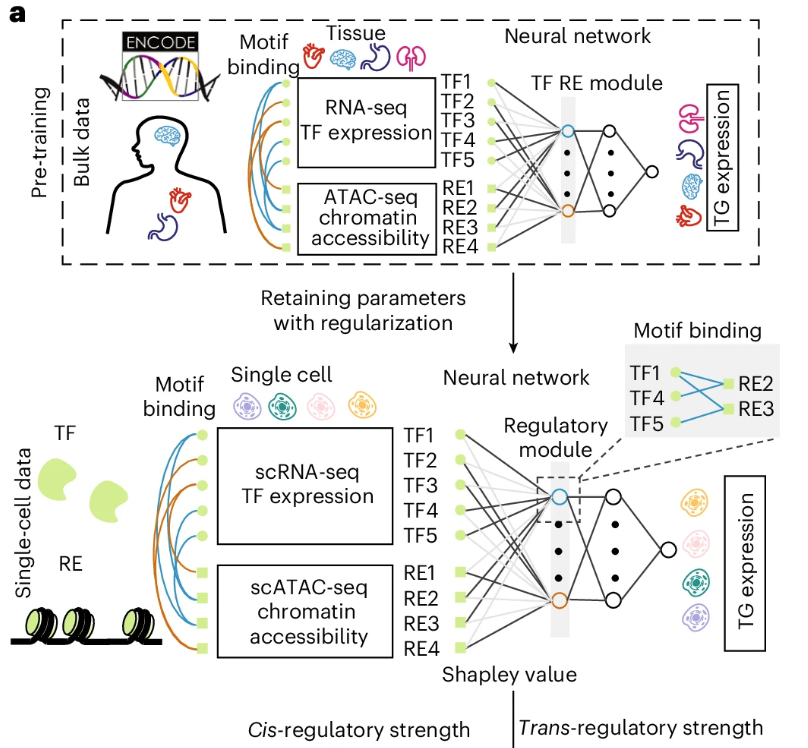
we present LINGER (Lifelong neural network for gene regulation), a machine-learning method to infer GRNs from single-cell paired gene expression and chromatin accessibility data. LINGER incorporates atlas-scale external bulk data across diverse cellular contexts and prior knowledge of transcription factor motifs as a manifold regularization.
Qiuyue Yuan and Zhana Duren
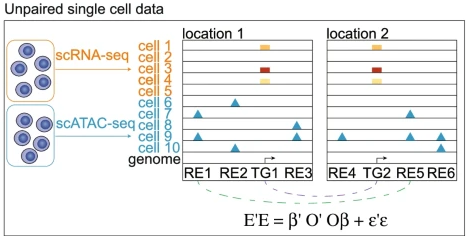
We propose a method named UnpairReg for the regression analysis on unpaired observations to integrate single-cell multi-omics data. On real and simulated data, UnpairReg provides an accurate estimation of cell gene expression where only chromatin accessibility data is available. The cis-regulatory network inferred from UnpairReg is highly consistent with eQTL mapping. UnpairReg improves cell type identification accuracy by joint analysis of single-cell gene expression and chromatin accessibility data.
Qiuyue Yuan and Zhana Duren
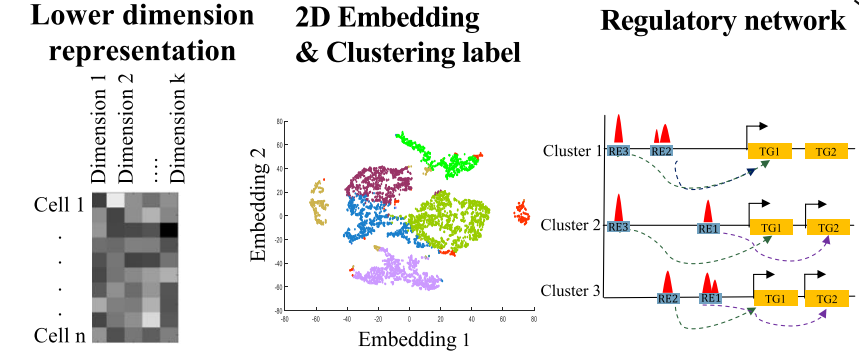
We develop scREG, a dimension reduction methodology, based on the concept of cis-regulatory potential, for single cell multiome data. This concept is further used for the construction of subpopulation-specific cis-regulatory networks. The R package scREG provides comprehensive functions for single cell multiome data analysis.
Zhana Duren, Fengge Chang, Fnu Naqing, Jingxue Xin, Qiao Liu, and Wing Hung Wong
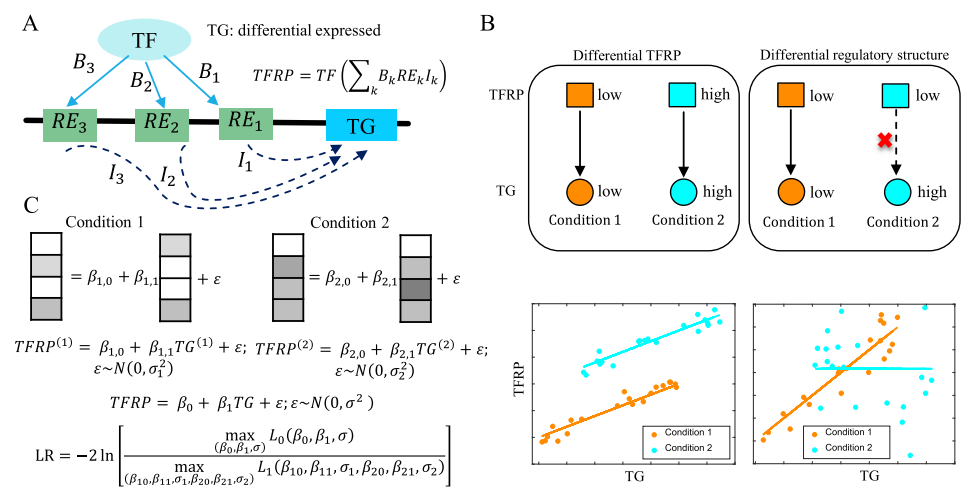
We propose sc-compReg as a method for the comparative analysis of gene expression regulatory networks between two conditions using single cell gene expression (scRNA-seq) and single cell chromatin accessibility data (scATAC-seq).
Zhana Duren, Sophia Lu, Joseph G. Arthur, Preyas Shah, Jingxue Xin, Francesca Meschi, Miranda Lin Li, Corey M. Nemec, Yifeng Yin, and Wing Hung Wong
Here we develop a Bayesian framework that integrates GWAS summary statistics with regulatory networks to infer genetic enrichments and associations simultaneously. Our method improves upon existing approaches by explicitly modeling network topology to assess enrichments, and by automatically leveraging enrichments to identify associations.
Xiang Zhu, Zhana Duren, Wing Hung Wong
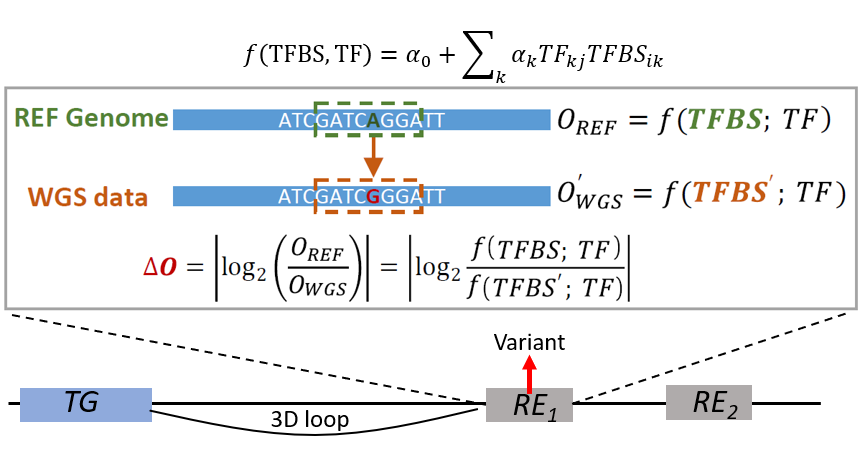
Here we use the expression and accessibility data from a diverse set of cell types to learn a model for the dependence of the accessibility of a regulatory element on its DNA sequence and TF expression. To interpret a personal genome, we combine the sequence information with context-specific TF expression to prioritize variants and regulatory elements in any genomic region of interest. This approach should be helpful in the study of risk loci previously identified by GWAS.
Wenran Li, Zhana Duren, Rui Jiang, Wing Hung Wong
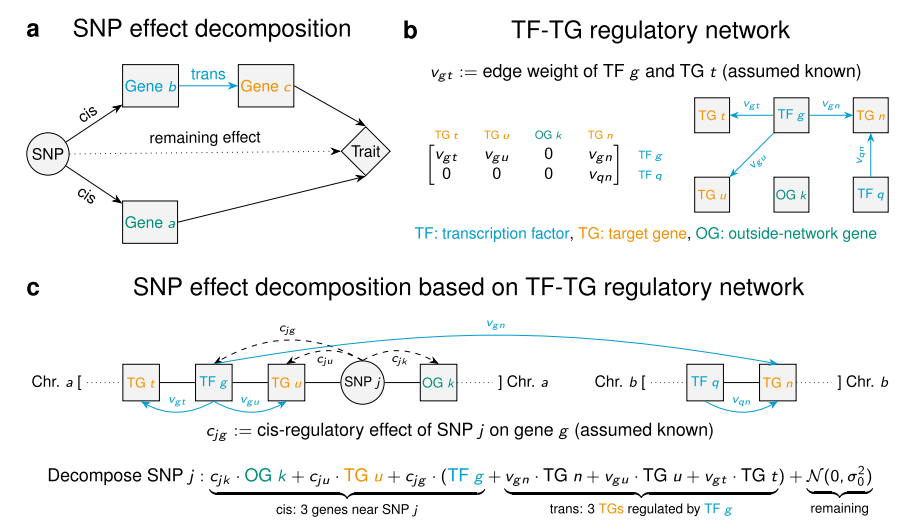
We develop a variant interpretation methodology (vPECA) to identify active selected REs (ASREs) and associated regulatory network. We discover three causal SNPs of EPAS1, the key adaptive gene for Tibetans. These SNPs decrease the accessibility of ASREs with weakened binding strength of relevant TFs, and cooperatively down-regulate EPAS1 expression.
Jingxue Xin, Hui Zhang, Yaoxi He, Zhana Duren, Caijuan Bai, Lang Chen, Xin Luo, Dong-Sheng Yan, Chaoyu Zhang, Xiang Zhu, Qiuyue Yuan, Zhanying Feng, Chaoying Cui, Xuebin Qi, Wing Hung Wong, Yong Wang, Bing Su
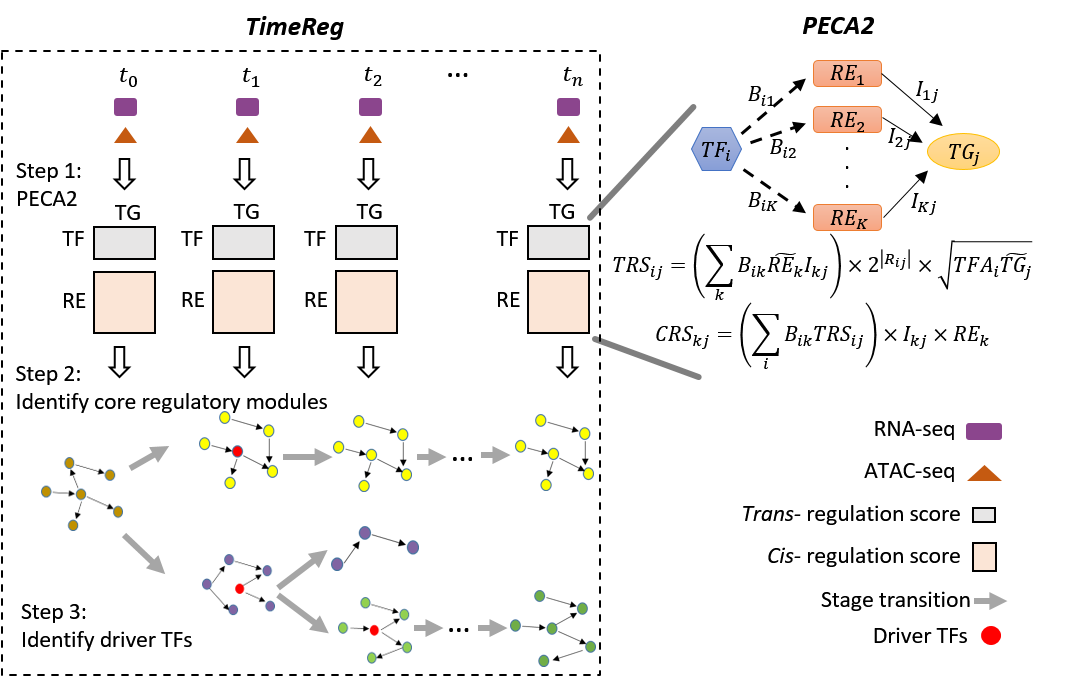
We develop a computational tool named TimeReg, which can be used to prioritize regulatory elements, to extract core regulatory modules at each time point, to identify key regulators driving changes of the cellular state, and to causally connect the modules across different time points.
Zhana Duren, Xi Chen, Jingxue Xin, Yong Wang, Wing Hung Wong
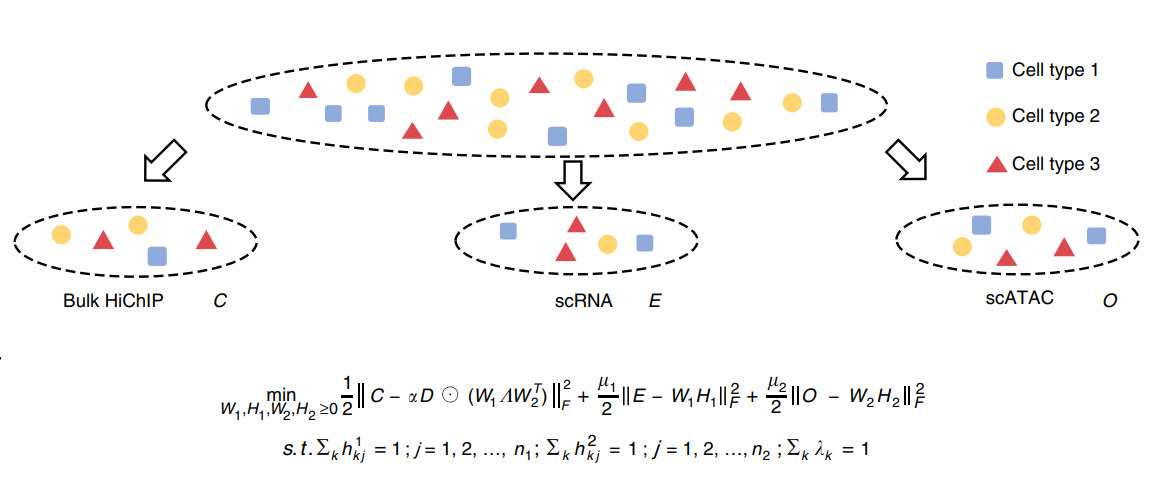
The first joint analysis method of bulk 3D chromatin interaction data with single-cell gene expression and chromatin accessibility from the same heterogeneous cell population. In this work, we introduce DC3 (De-Convolution and Coupled-Clustering) as a method for the joint analysis of various bulk and single-cell data such as HiChIP, RNA-seq and ATAC-seq from the same heterogeneous cell population. DC3 can simultaneously identify distinct subpopulations, assign single cells to the subpopulations (i.e., clustering) and de-convolve the bulk data into subpopulation-specific data.
Wanwen Zeng, Xi Chen, Zhana Duren, Yong Wang, Rui Jiang, Wing Hung Wong
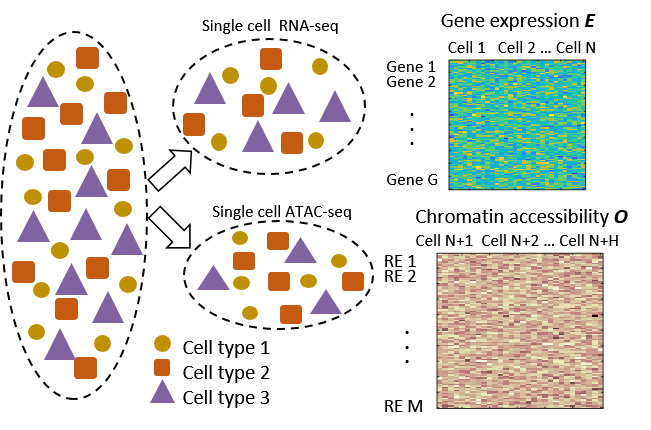
Biological samples are often heterogeneous mixtures of different types of cells. Suppose we have two single-cell datasets, each providing information on a different cellular feature and generated on a different sample from this mixture. Then, the clustering of cells in the two samples should be coupled as both clusterings are reflecting the underlying cell types in the same mixture. This “coupled clustering” problem is a new problem not covered by existing clustering methods. In this paper, we develop an approach for its solution based on the coupling of two nonnegative matrix factorizations. The method should be useful for integrative single-cell genomics analysis tasks such as the joint analysis of single-cell RNA-sequencing and single-cell ATAC-sequencing data.
Zhana Duren, Xi Chen, Mahdi Zamanighomi, Wanwen Zeng, Ansuman T. Satpathy, Howard Y. Chang, Yong Wang, Wing Hung Wong
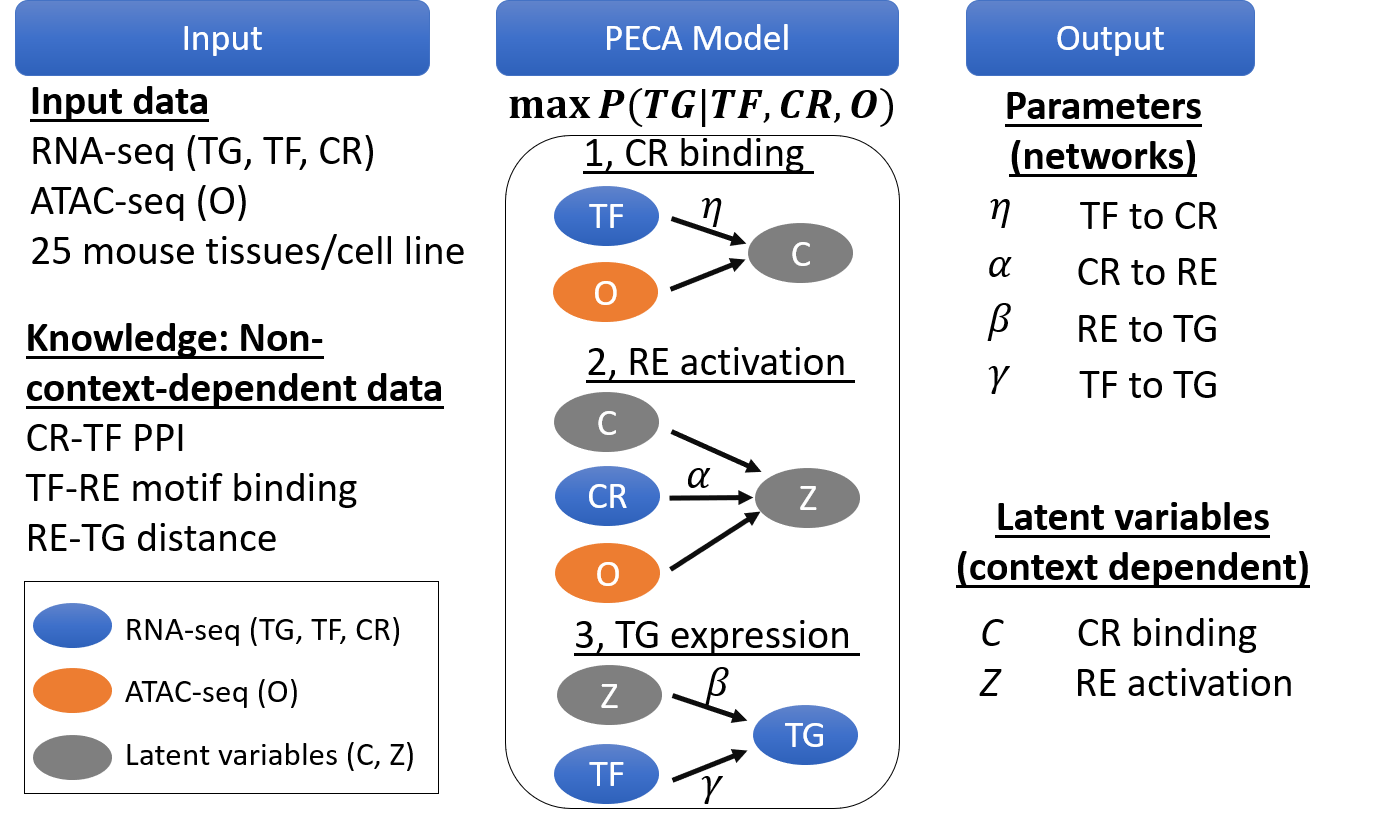
The PECA is a computational mechanistic model for gene regulatory network from paired gene expression and chroamtin accessibility data. Chromatin plays a critical role in the regulation of gene expression. Interactions among chromatin regulators, sequence-specific transcription factors, and cis-regulatory sequence elements are the main driving forces shaping context-specific chromatin structure and gene expression. The purpose of the present work is to show that, by modeling matched expression and accessibility data across diverse cellular contexts, it is possible to recover a significant portion of the information in the missing data on binding locations and chromatin states and to achieve accurate inference of gene regulatory relations.
Zhana Duren, Xi Chen, Rui Jiang, Yong Wang, Wing Hung Wong
Full List
Inferring gene regulatory networks from single-cell multiome data using atlas-scale external data
Qiuyue Yuan and Zhana Duren
Nature Biotechnology 2024
Joint inference of clonal structure using single-cell genome and transcriptome sequencing data
Xiangqi Bai, Zhana Duren, Lin Wan, Li C Xia
NAR Genomics and Bioinformatics 2024
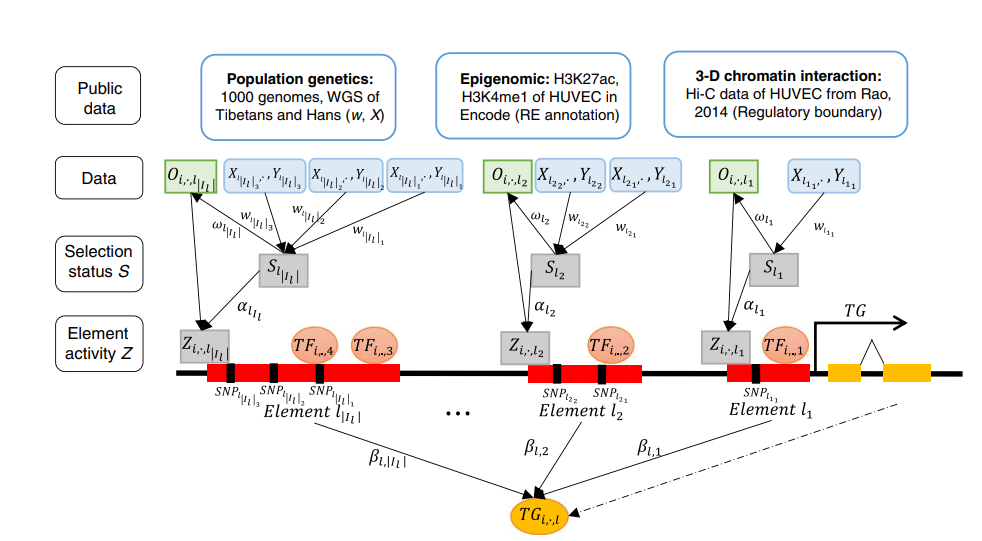
Heritability enrichment in context-specific regulatory networks improves phenotype-relevant tissue identification
Zhanying Feng, Zhana Duren, Jingxue Xin, Qiuyue Yuan , Yaoxi He, Bing Su, Wing Hung Wong, and Yong Wang
eLife 2022
Human genetic variants associated with COVID-19 severity are enriched in immune and epithelium regulatory networks
Zhanying Feng, Xianwen Ren, Zhana Duren, and Yong Wang
Phenomics 2022
Integration of single-cell multi-omics data by regression analysis on unpaired observations
Qiuyue Yuan and Zhana Duren
Genome Biology 2022
Regulatory analysis of single cell multiome gene expression and chromatin accessibility data with scREG
Zhana Duren, Fengge Chang, Fnu Naqing, Jingxue Xin, Qiao Liu, and Wing Hung Wong
Genome Biology 2022
Sc-compReg enables the comparison of gene regulatory networks between conditions using single-cell data
Zhana Duren, Sophia Lu, Joseph G. Arthur, Preyas Shah, Jingxue Xin, Francesca Meschi, Miranda Lin Li, Corey M. Nemec, Yifeng Yin, and Wing Hung Wong
Nature communications 2021
Dynamic chromatin regulatory landscape of human CAR T cell exhaustion
David G. Gennert, Rachel C. Lynn, Jeff M. Granja, Evan W. Weber, Maxwell R. Mumbach, Yang Zhao, Zhana Duren, Elena Sotillo, William J. Greenleaf, Wing H. Wong, Ansuman T. Satpathy, Crystal L. Mackall, and Howard Y. Chang
PNAS 2021
MIMIC: an optimization method to identify cell type-specific marker panel for cell sorting
Meng Zou, Zhana Duren, Qiuyue Yuan, Henry Li, Andrew Paul Hutchins, Wing Hung Wong, Yong Wang
Briefings in Bioinformatics 2021
hReg-CNCC reconstructs a regulatory network in human cranial neural crest cells and annotates variants in a developmental context
Zhanying Feng, Zhana Duren, Ziyi Xiong, Sijia Wang, Fan Liu, Wing Hung Wong, Yong Wang
Communications biology 2021
Modeling regulatory network topology improves genome-wide analyses of complex human traits
Xiang Zhu, Zhana Duren, Wing Hung Wong
Nature communications 2021
GuidingNet: revealing transcriptional cofactor and predicting binding for DNA methyltransferase by network regularization
Lixin Ren, Caixia Gao, Zhana Duren, Yong Wang
Briefings in Bioinformatics 2021
A method for scoring the cell type-specific impacts of noncoding variants in personal genomes
Wenran Li, Zhana Duren, Rui Jiang, Wing Hung Wong
PNAS 2020
Chromatin accessibility landscape and regulatory network of high-altitude hypoxia adaptation
Jingxue Xin, Hui Zhang, Yaoxi He, Zhana Duren, Caijuan Bai, Lang Chen, Xin Luo, Dong-Sheng Yan, Chaoyu Zhang, Xiang Zhu, Qiuyue Yuan, Zhanying Feng, Chaoying Cui, Xuebin Qi, Wing Hung Wong, Yong Wang, Bing Su
Nature communications 2020
Time course regulatory analysis based on paired expression and chromatin accessibility data
Zhana Duren, Xi Chen, Jingxue Xin, Yong Wang, Wing Hung Wong
Genome Research 2020
Integrated functional genomic analyses of Klinefelter and Turner syndromes reveal global network effects of altered X chromosome dosage
Xianglong Zhang, David Hong, Shining Ma, Thomas Ward, Marcus Ho, Reenal Pattni, Zhana Duren, Atanas Stankov, Sharon Bade Shrestha, Joachim Hallmayer, Wing Hung Wong, Allan L Reiss, Alexander E Urban
PNAS 2020
Modeling regulatory network topology improves genome-wide analyses of complex human traits
Xiang Zhu, Zhana Duren, Wing Hung Wong
bioRxiv 2020
DC3 is a method for deconvolution and coupled clustering from bulk and single-cell genomics data
Wanwen Zeng, Xi Chen, Zhana Duren, Yong Wang, Rui Jiang, Wing Hung Wong
Nature Communications 2019
Hierarchical graphical model reveals HFR1 bridging circadian rhythm and flower development in Arabidopsis thaliana
Zhana Duren, Yaling Wang, Jiguang Wang, Xing-Ming Zhao, Le Lv, Xiaobo Li, Jingdong Liu, Xin-Guang Zhu, Luonan Chen, Yong Wang
NPJ systems biology and applications 2019
TFAP2C-and p63-dependent networks sequentially rearrange chromatin landscapes to drive human epidermal lineage commitment
Lingjie Li, Yong Wang, Jessica L Torkelson, Gautam Shankar, Jillian M Pattison, Hanson H Zhen, Fengqin Fang, Zhana Duren, Jingxue Xin, Sadhana Gaddam, Sandra P Melo, Samantha N Piekos, Jiang Li, Eric J Liaw, Lang Chen, Rui Li, Marius Wernig, Wing H Wong, Howard Y Chang, Anthony E Oro
Cell Stem Cell 2019
Integrative analysis of single-cell genomics data by coupled nonnegative matrix factorizations
Zhana Duren, Xi Chen, Mahdi Zamanighomi, Wanwen Zeng, Ansuman T. Satpathy, Howard Y. Chang, Yong Wang, Wing Hung Wong
PNAS 2018
Unsupervised clustering and epigenetic classification of single cells
Mahdi Zamanighomi, Zhixiang Lin, Timothy Daley, Xi Chen, Zhana Duren, Alicia Schep, William J Greenleaf, Wing Hung Wong
Nature communications 2018
Modeling gene regulation from paired expression and chromatin accessibility data
Zhana Duren, Xi Chen, Rui Jiang, Yong Wang, Wing Hung Wong
PNAS 2017
A systematic method to identify modulation of transcriptional regulation via chromatin activity reveals regulatory network during mESC differentiation
Zhana Duren, Yong Wang
Scientific reports 2016
A community effort to assess and improve drug sensitivity prediction algorithms
James C Costello, Laura M Heiser, Elisabeth Georgii et. al
Nature biotechnology 2014
Sparse hyperspectral unmixing using an approximate L0 norm
Wei Tang, Zhenwei Shi, Zhana Duren
Optik 2014
Subspace matching pursuit for sparse unmixing of hyperspectral data
Zhenwei Shi, Wei Tang, Zhana Duren, Zhiguo Jiang
IEEE Transactions on Geoscience and Remote Sensing 2013
Inferring gene regulatory network for cell reprogramming
Duren Zhana , Wang Yong, Saito Shigeru, Horimoto Katsuhisa
Control Conference (CCC), 2012 31st Chinese
Clinical data analysis reveals three subytpes of gastric cancer
Xinxin Wang, Zhana Duren, Chao Zhang, Lin Chen, Yong Wang
2012 IEEE 6th International Conference on Systems Biology (ISB)