Welcome to the Duren Lab
We are a computational biology research group at the Department of Medical and Molecular Genetics and Center for Computational Biology and Bioinformatics of Indiana University School of Medicine. Our aim is to explore and understand the regulatory mechanisms of non-coding DNA. The long-term research goal is to answer the key scientific question” How non-coding genetic variant act through context specific gene regulatory network to influence phenotype”.
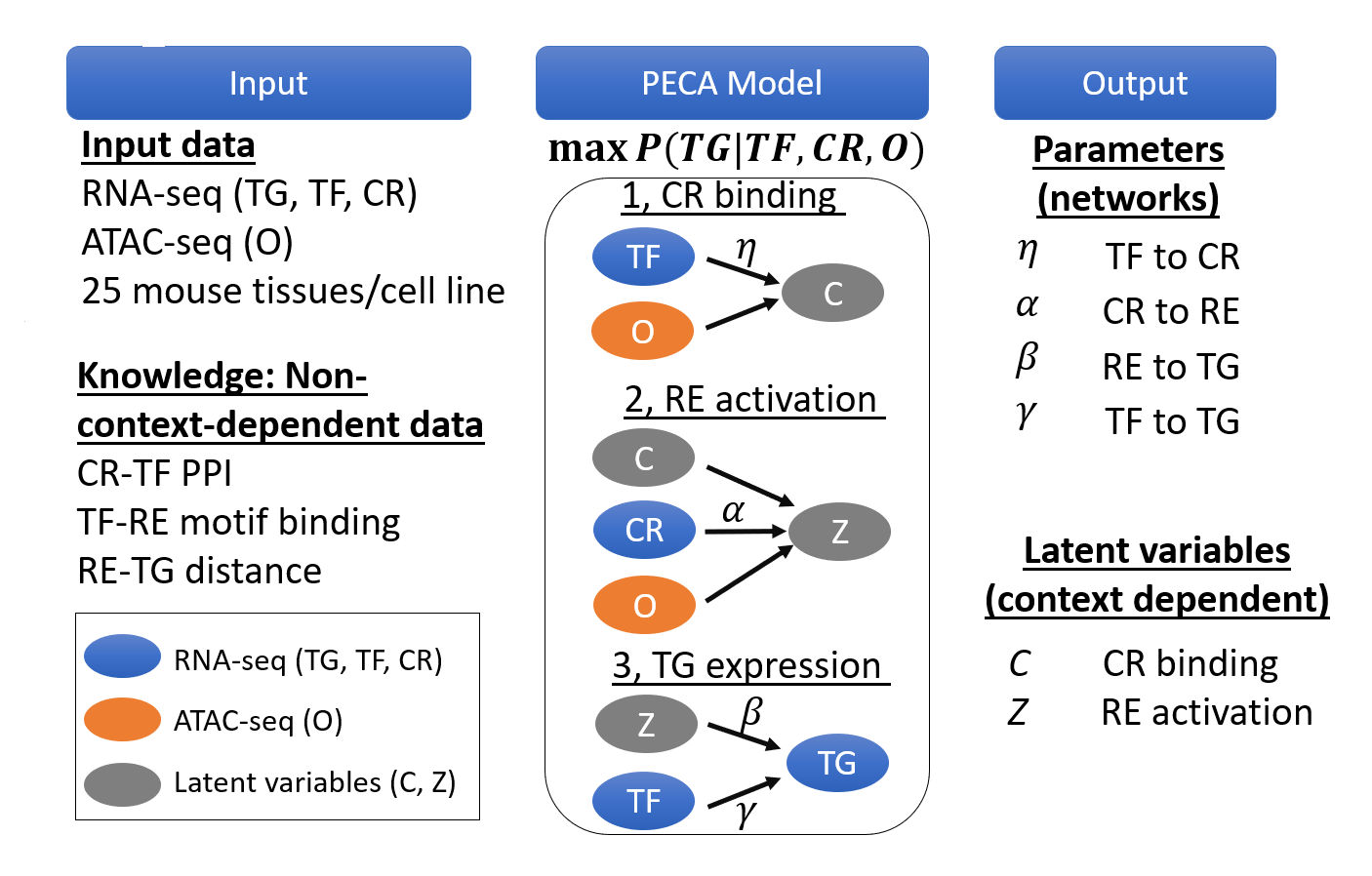
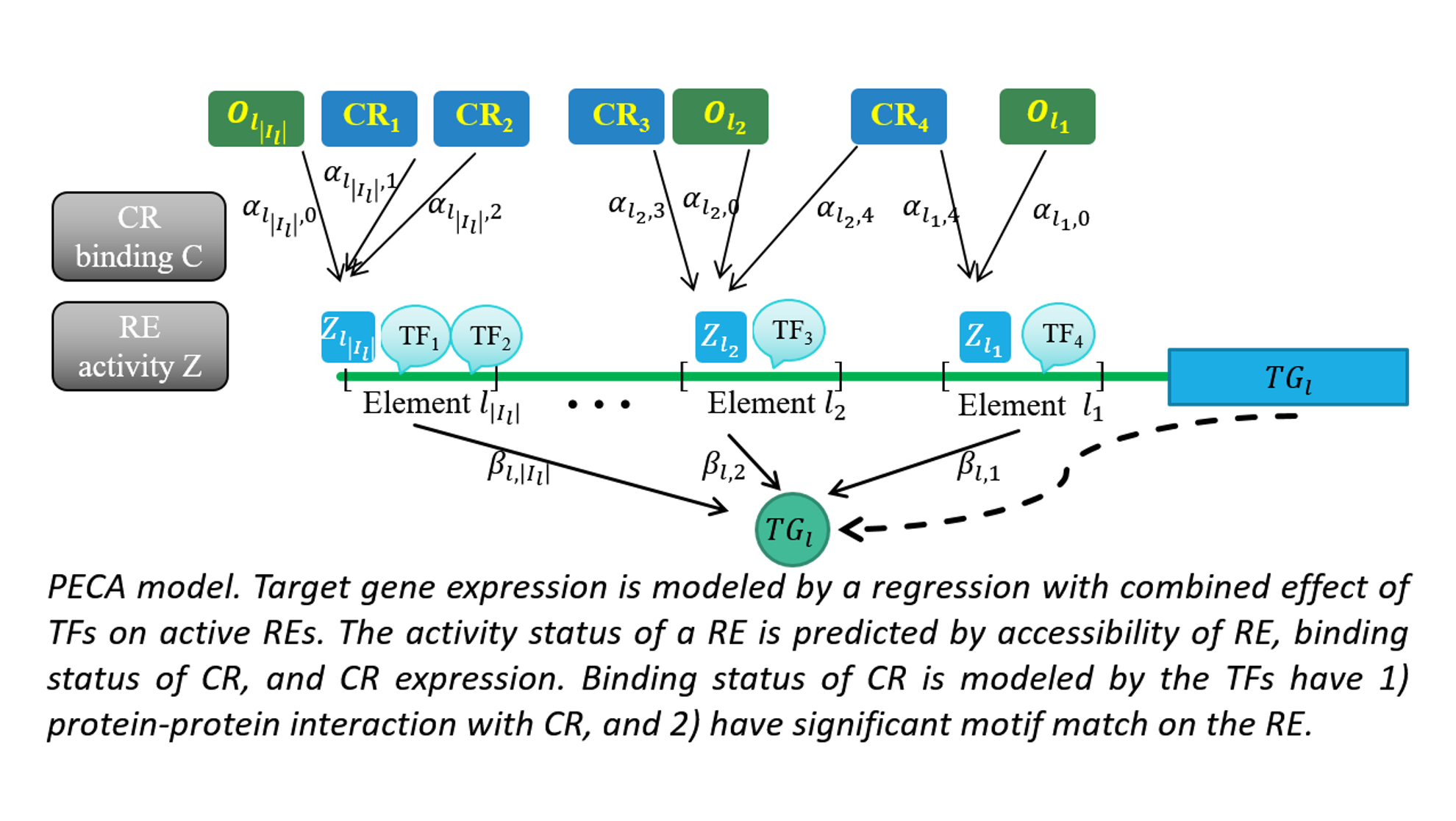
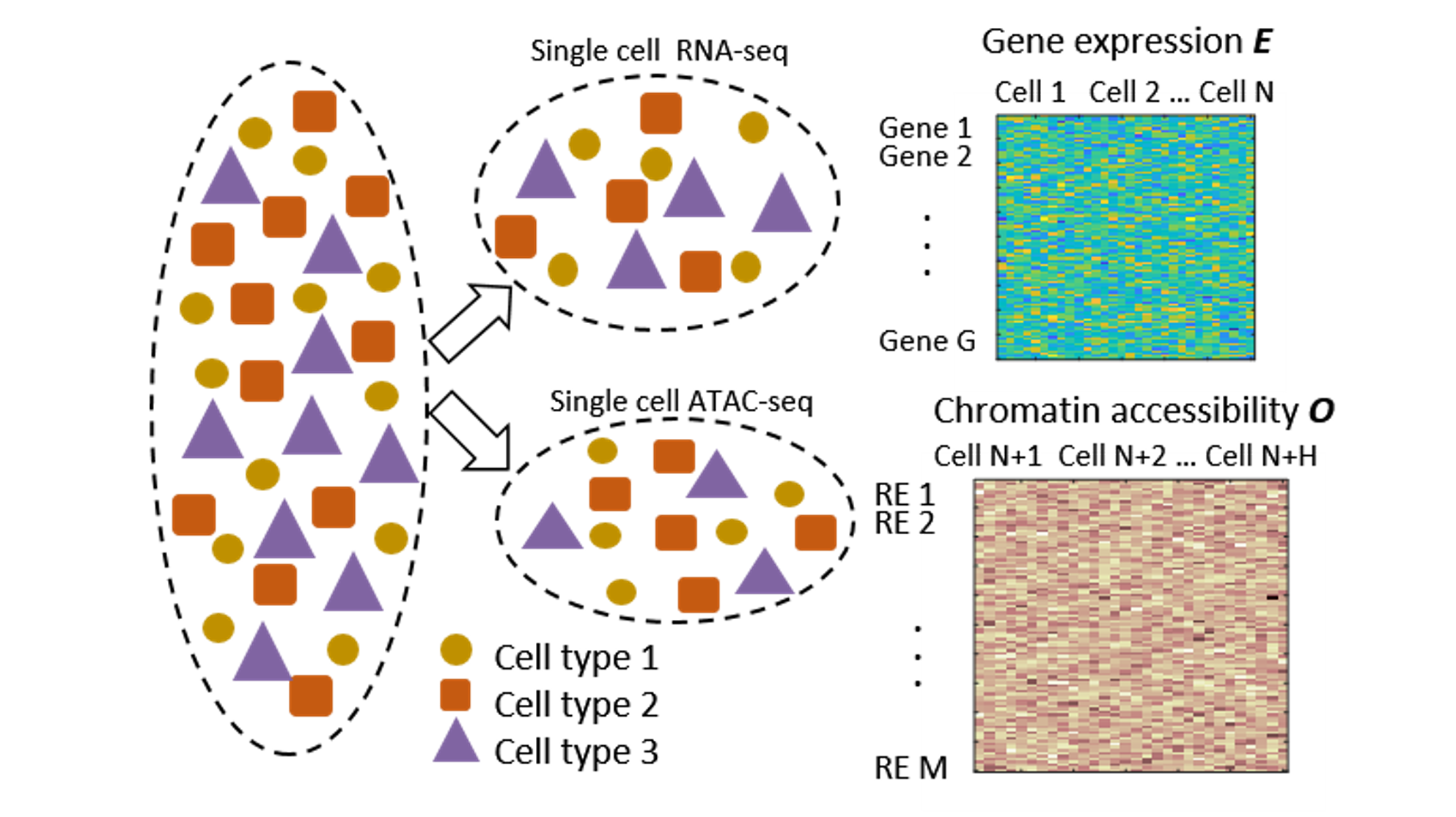
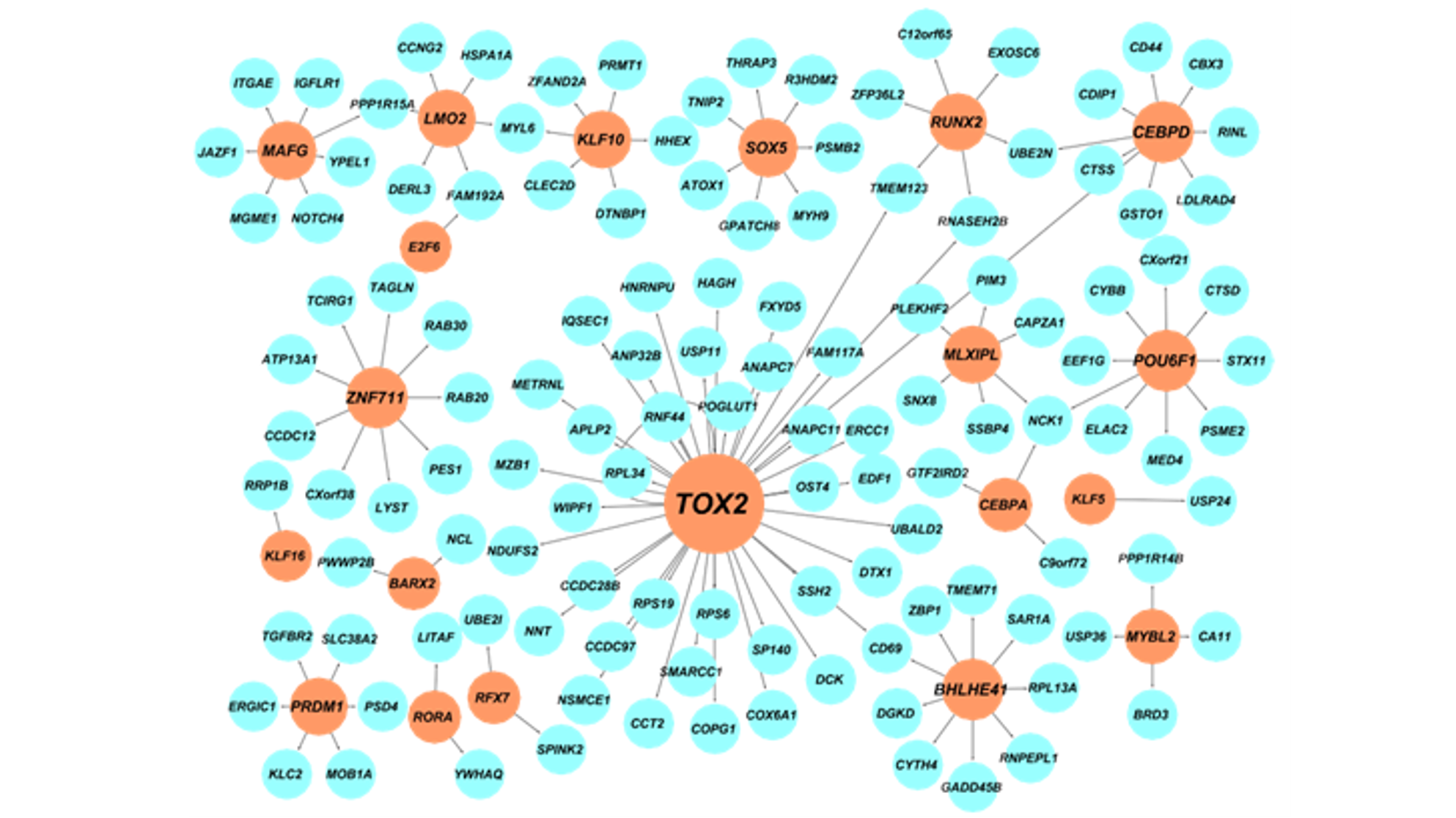
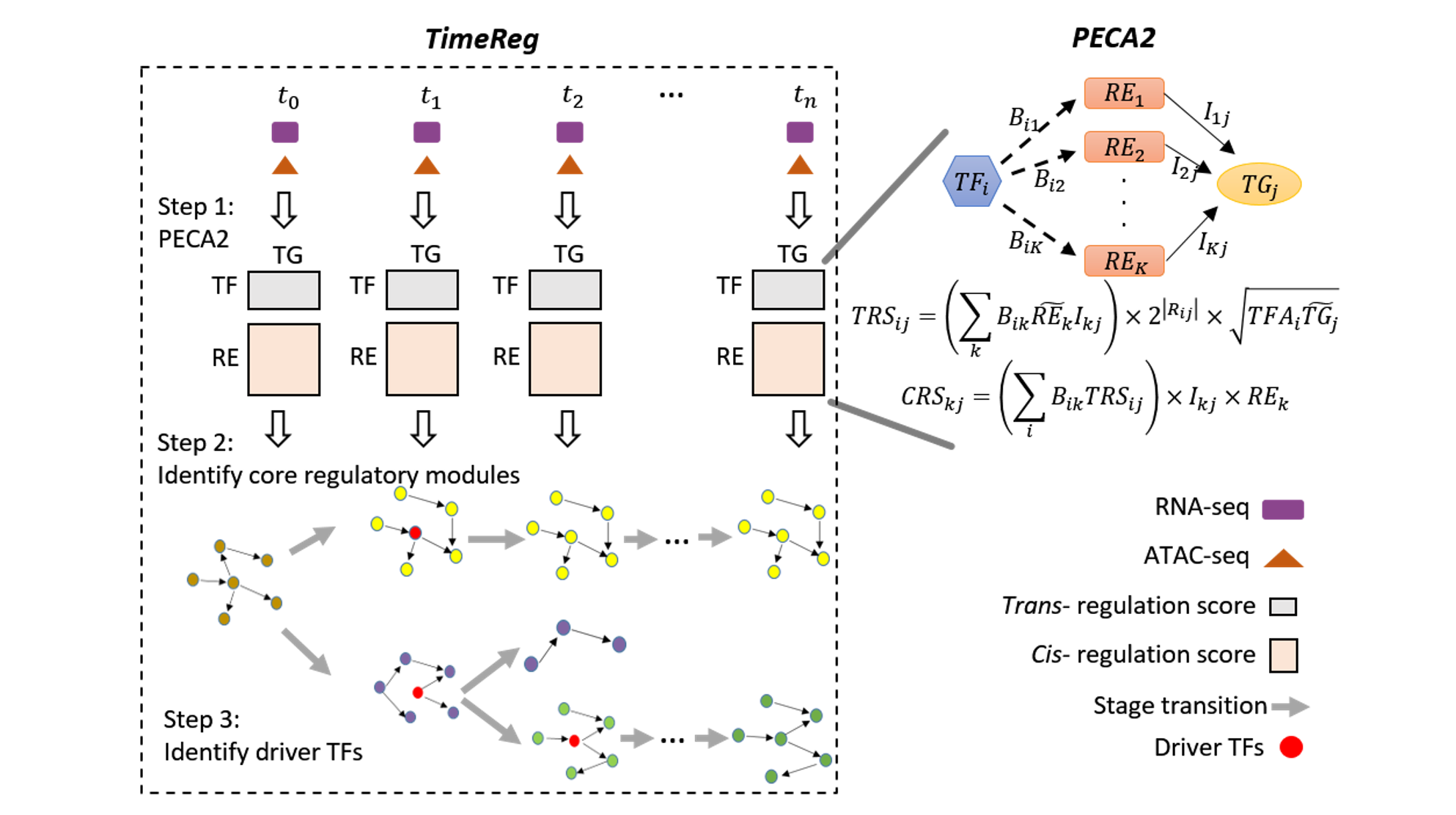
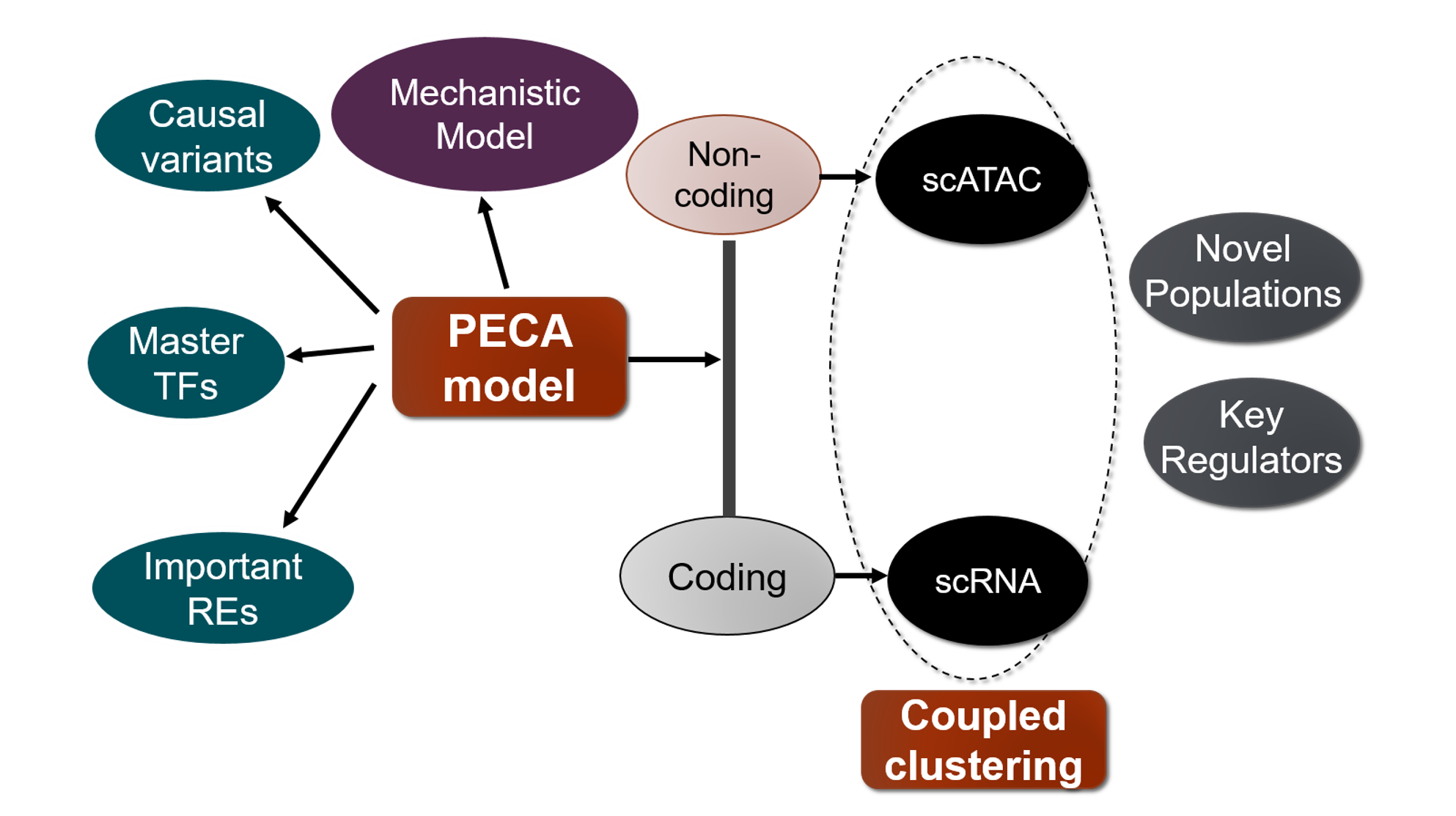
To this end, we develop novel computational tools for integrative analysis of genomics data to infer gene regulatory networks, to detect heterogeneity, and to annotate the regulatory function of genetic variants by context specific manner. (see Research).
We are part of the Indiana University School of Medicine Department of Medical and Molecular Genetics, which is located in 410 W. 10th Street, a state-of-the-art research and educational facility located in Indianapolis, Indiana.
We are looking for new Postdocs, graduate students, and ungraduate students to join the team (more info) !
News
25 August 2025Graduate student Su Xu joined our lab!
1 November 2024Our lab moved to Indiana University.
12 April 2024The Linger is published on Nature Biotechnology . Congrats Qiuyue.
12 April 2024We are reported on Clemson News , Go Tigers!
29 March 2024Zhana visited the Center for Computational Biology and Bioinformatics at the Indiana University and presented the LINGER method at the seminar.
22 March 2024We attended the MCBIOS2024 conference, which Zhana co-organized. Zhana and Ishita gave talk presentations. Naqing and Hui presented posters.
15 March 2024We recived 2 Years R21 fund from NIDA/NIH, direct cost $275K!
15 March 2024Zhana served on the review panel of data integration and statistical analysis methods for dGTEx!
1 Feb 2024Zhana served as a councilors for NIGMS/NIH concil meeting.